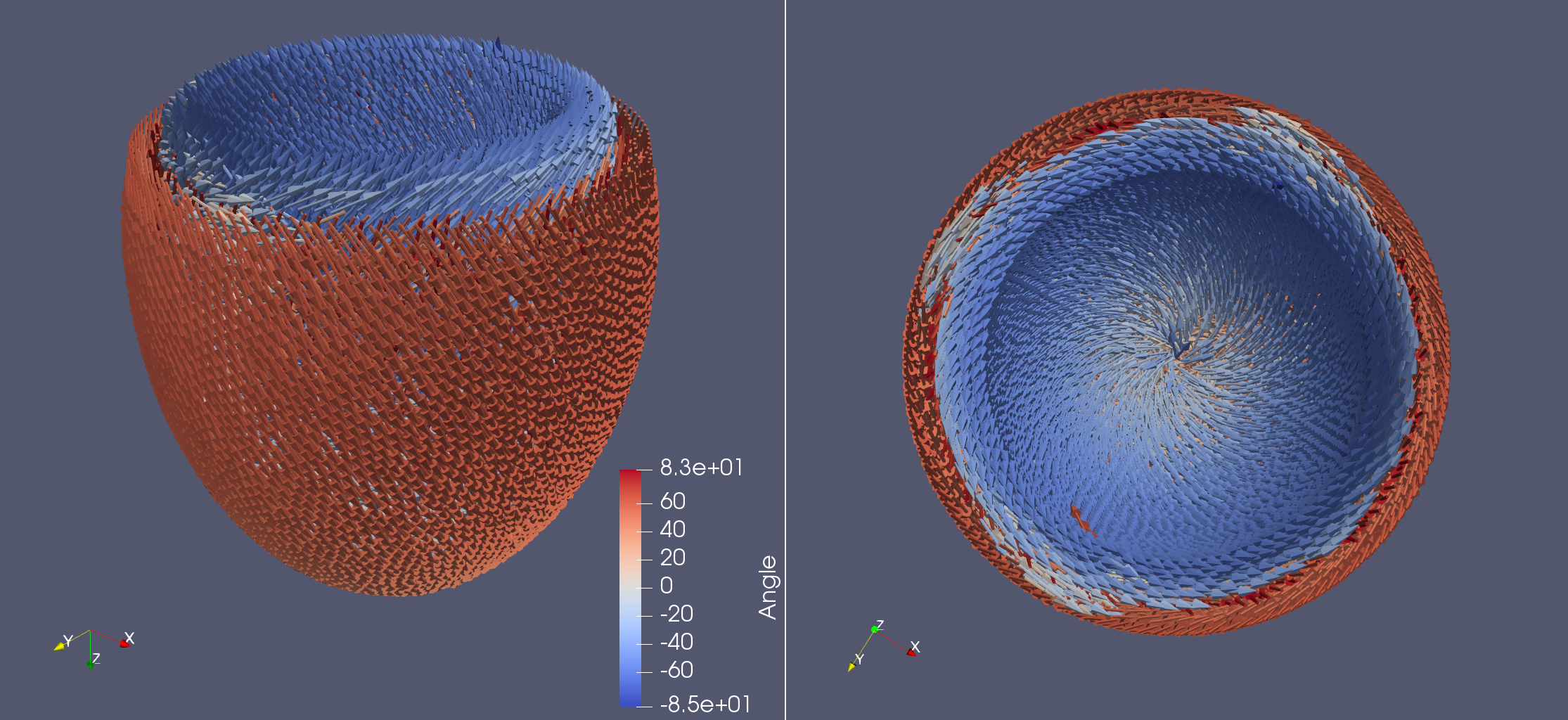
Life X - idealized_LV#
Life X recently published their own implementation of the LDRB algorithm and with that they also published a lot of example meshes. This demo aims to try out this implementation of the LDRB algorithm on these example meshes. The LifeX example meshes can be found at https://zenodo.org/record/5810269#.YeEjWi8w1B0, which also contains a DOI: https://doi.org/10.5281/zenodo.5810269.
This demo assumes that you have downloaded the folder with the meshes in the same format as they are uploaded on zenodo, so that the gmsh files are located in a folder called lifex_fiber_generation_examples/mesh.
First we import the necessary packages. Note that we also import meshio which is used for converted from .msh (gmsh) to .xdmf (FEnICS).
import dolfin
import ldrb
import cardiac_geometries
Load the mesh and markers.
mesh, markers, marker_functions = cardiac_geometries.gmsh2dolfin(
"lifex_fiber_generation_examples/mesh/idealized_LV.msh",
)
Here the markers are actually parsed with meshio, but they have the wrong name so we just rename them
ldrb_markers = {
"epi": markers["Epicardium"][0],
"lv": markers["Endocardium"][0],
"base": markers["Basal plane"][0],
}
Select linear Lagrange elements
fiber_space = "P_1"
Compute the fiber-sheet system
fiber, sheet, sheet_normal = ldrb.dolfin_ldrb(
mesh=mesh,
fiber_space=fiber_space,
ffun=marker_functions.ffun,
markers=ldrb_markers,
alpha_endo_lv=60, # Fiber angle on the endocardium
alpha_epi_lv=-60, # Fiber angle on the epicardium
beta_endo_lv=0, # Sheet angle on the endocardium
beta_epi_lv=0, # Sheet angle on the epicardium
)
And save the results
with dolfin.XDMFFile(mesh.mpi_comm(), "idealized_LV_fiber.xdmf") as xdmf:
xdmf.write(fiber)