Inducing reentry in a 2D sheet of cardiac tissue#
In this demo we show how to induce reentry in a 2D sheet of cardiac tissue using two stimulations, i.e one wave where we stimulate the left side of the square and then one second stimulation where we stimulate the third quadrant of the square.
First we make the necessary imports
from pathlib import Path
import dolfin
import matplotlib.pyplot as plt
import numpy as np
import gotranx
import beat
And set up some basic parameters
results_folder = Path("results-reentry")
save_every_ms = 1.0
transverse = False
# Increase this to make the simulation longer
end_time = 10.0
dt = 0.05
save_freq = round(save_every_ms / dt)
overwrite = True
mesh_unit = "cm"
dx = 0.4 * beat.units.ureg("mm").to(mesh_unit).magnitude
L = 5.0 * beat.units.ureg("cm").to(mesh_unit).magnitude
data = beat.geometry.get_2D_slab_geometry(
Lx=L,
Ly=L,
dx=dx,
transverse=transverse,
)
V = dolfin.FunctionSpace(data.mesh, "CG", 1)
For this simulation we will use a model from [CRN98]. The model is taken form https://models.physiomeproject.org/workspace/courtemanche_ramirez_nattel_1998
print("Running model")
Running model
Load the model
model_path = Path("courtemanche_ramirez_nattel_1998.py")
if not model_path.is_file():
here = Path.cwd()
ode = gotranx.load_ode(
here / ".." / "odes" / "courtemanche_ramirez_nattel_1998.ode"
)
code = gotranx.cli.gotran2py.get_code(
ode, scheme=[gotranx.schemes.Scheme.forward_generalized_rush_larsen]
)
model_path.write_text(code)
import courtemanche_ramirez_nattel_1998
model = courtemanche_ramirez_nattel_1998.__dict__
Specify the membrane capacitance and the surface to volume ratio
C_m = 1.0 * beat.units.ureg("uF/cm**2")
chi = 1000.0 * beat.units.ureg("cm**-1")
Here we also reduce the conductance for the sodium channel to make sure that the speed of the traveling wave is reduced
fun = model["forward_generalized_rush_larsen"]
y = model["init_state_values"]()
time = dolfin.Constant(0.0)
parameters = model["init_parameter_values"](stim_amplitude=0.0)
The duration of the stimulation is set to 5 ms
duration = 5.0
We don’t know when the S2 stimulation should be triggered, and we will find this by monitoring the voltage in the S2 domain. For now let us just create a placeholder for the start of the S2 stimulation and just set the value to a large number
s2_start = dolfin.Constant(9999999.0)
We specify this stimulation protocol as an expressions. We will first stimulate the left boundary and then after a delay we will stimulate the lower left quadrant. In this case we can very easily define this in a single expression as
expr_single_s1s2 = dolfin.Expression(
"(t < duration && x[0] < 0.05) ? 20.0 : "
"((t > delay && t < delay+duration) && (x[0] < L / 2 && x[1] < L / 2)) ? 20.0 : 0.0",
t=time,
L=L,
delay=s2_start,
duration=duration,
degree=0,
)
and then define the stimulus as
I_s_s1s2 = beat.base_model.Stimulus(
dz=dolfin.dx, expr=expr_single_s1s2
)
However, sometimes you might have a mesh function defining the stimulus domains, in which case it can be hard to do this using a single expression. Therefore we will show an approach where we have two different expressions and two different currents representing the two stimuli.
First we define the S1 stimulation
s1s2_markers = dolfin.MeshFunction("size_t", data.mesh, 2)
s1s2_markers.set_all(0)
s1_domain = dolfin.CompiledSubDomain("x[0] < 0.05")
s1_marker = 1
s1_domain.mark(s1s2_markers, s1_marker)
I_s_s1 = beat.stimulation.define_stimulus(
chi=chi,
mesh=data.mesh,
mesh_unit=mesh_unit,
time=time,
start=0.0,
duration=duration,
amplitude=20_000,
subdomain_data=s1s2_markers,
marker=s1_marker
)
s2_domain = dolfin.CompiledSubDomain("x[0] > 0.05 && x[0] < L / 2 && x[1] < L / 2", L=L)
s2_marker = 2
s2_domain.mark(s1s2_markers, s2_marker)
I_s_s2 = beat.stimulation.define_stimulus(
chi=chi,
mesh=data.mesh,
mesh_unit=mesh_unit,
time=time,
start=s2_start,
duration=duration,
amplitude=20_000,
subdomain_data=s1s2_markers,
marker=s2_marker
)
Let us also compute the volume (or area) of the S2 domain which we will use later
vol_s2_domain = dolfin.assemble(dolfin.Constant(1) * I_s_s2.dz)
Calling FFC just-in-time (JIT) compiler, this may take some time.
Finally let us collect the two simulation currents into a list
I_s = [I_s_s1, I_s_s2]
Next we set up the models
V_ode = dolfin.FunctionSpace(data.mesh, "Lagrange", 1)
M = beat.conductivities.define_conductivity_tensor(
f0=data.f0,
**beat.conductivities.default_conductivities("Niederer"),
)
pde = beat.MonodomainModel(time=time, mesh=data.mesh, M=M, I_s=I_s, C_m=C_m.magnitude)
ode = beat.odesolver.DolfinODESolver(
v_ode=dolfin.Function(V_ode),
v_pde=pde.state,
fun=fun,
init_states=y,
parameters=parameters,
num_states=len(y),
v_index=model["state_index"]("V"),
)
solver = beat.MonodomainSplittingSolver(pde=pde, ode=ode)
fname = (results_folder / "V.xdmf").as_posix()
beat.postprocess.delete_old_file(fname)
Cannot determine geometric dimension from expression.
Calling FFC just-in-time (JIT) compiler, this may take some time.
Calling FFC just-in-time (JIT) compiler, this may take some time.
And create a function to be called when saving
def save(t):
v = solver.pde.state.vector().get_local()
print(f"Solve for {t=:.2f}, {v.max() =}, {v.min() = }")
with dolfin.XDMFFile(data.mesh.mpi_comm(), fname) as xdmf:
xdmf.parameters["functions_share_mesh"] = True
xdmf.parameters["rewrite_function_mesh"] = False
xdmf.write_checkpoint(
solver.pde.state,
"V",
float(t),
dolfin.XDMFFile.Encoding.HDF5,
True,
)
Finally we solve it. Here we also compute the average voltage inside the S2 domain, and if time is lager than 100 ms and the voltage drops below - 70 mV we will initiate the S2 stimulus
t = 0.0
save_freq = int(1.0 / dt)
i = 0
s2_triggered = False
while t < end_time + 1e-12:
# Make sure to save at the same time steps that is used by Ambit
if i % save_freq == 0:
save(t)
# Check if average voltage in S2 domain is below threshold
v_s2_avg = dolfin.assemble(solver.pde.state * I_s_s2.dz) / vol_s2_domain
print(f"Average voltage in S2 domain = {v_s2_avg}")
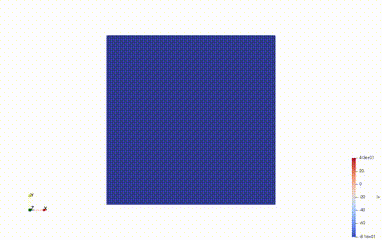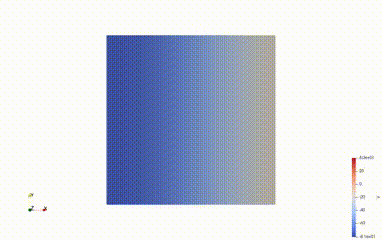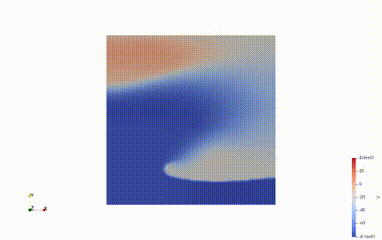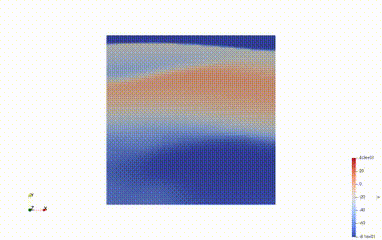
if not s2_triggered and (t > 100.0 and v_s2_avg < -70.0):
print("Initiating S2 stimulation")
s2_start.assign(t)
s2_triggered = True
solver.step((t, t + dt))
i += 1
t += dt
Solve for t=0.00, v.max() =-81.17999999999999, v.min() = -81.18000000000008
Calling FFC just-in-time (JIT) compiler, this may take some time.
Average voltage in S2 domain = -81.1800000000204
Calling FFC just-in-time (JIT) compiler, this may take some time.
Solve for t=1.00, v.max() =-63.38588556115019, v.min() = -81.54036238307688
Average voltage in S2 domain = -81.1698480158202
Solve for t=2.00, v.max() =-51.25235261154096, v.min() = -81.35679992662975
Average voltage in S2 domain = -81.11378511346608
Solve for t=3.00, v.max() =7.481268708218495, v.min() = -81.28134774704493
Average voltage in S2 domain = -81.0271782034219
Solve for t=4.00, v.max() =27.527489081791433, v.min() = -81.97653946114792
Average voltage in S2 domain = -80.3792672733849
Solve for t=5.00, v.max() =28.480568242169774, v.min() = -82.43576149908489
Average voltage in S2 domain = -78.5448337510221
Solve for t=6.00, v.max() =11.79832459449342, v.min() = -82.58183112713155
Average voltage in S2 domain = -76.89778643012282
Solve for t=7.00, v.max() =9.757449407801051, v.min() = -82.91801804849592
Average voltage in S2 domain = -75.39893085243143
Solve for t=8.00, v.max() =8.003862849056112, v.min() = -82.87134503727198
Average voltage in S2 domain = -73.71706959733662
Solve for t=9.00, v.max() =7.148887152709796, v.min() = -82.08527380658997
Average voltage in S2 domain = -71.82918811743095
Solve for t=10.00, v.max() =6.899651388236039, v.min() = -82.46304112534533
Average voltage in S2 domain = -70.30942579619904
Marc Courtemanche, Rafael J Ramirez, and Stanley Nattel. Ionic mechanisms underlying human atrial action potential properties: insights from a mathematical model. American Journal of Physiology-Heart and Circulatory Physiology, 275(1):H301–H321, 1998.
Sander Land, Viatcheslav Gurev, Sander Arens, Christoph M Augustin, Lukas Baron, Robert Blake, Chris Bradley, Sebastian Castro, Andrew Crozier, Marco Favino, and others. Verification of cardiac mechanics software: benchmark problems and solutions for testing active and passive material behaviour. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences, 471(2184):20150641, 2015.
Zhaoyang Zhang, Michael B Liu, Xiaodong Huang, Zhen Song, and Zhilin Qu. Mechanisms of premature ventricular complexes caused by qt prolongation. Biophysical journal, 120(2):352–369, 2021.
