Conduction velocity and ECG for slabs#
In this demo we will show how to compute conduction velocity and ECG for a Slab geometry.
from pathlib import Path
import beat.conductivities
import beat.postprocess
import beat.single_cell
import beat.stimulation
import dolfin
import matplotlib.pyplot as plt
import numpy as np
import gotranx
import beat
import pyvista
import beat.viz
from beat.geometry import Geometry
results_folder = Path("results-slab")
save_every_ms = 1.0
dimension = 2
transverse = False
end_time = 20.0
dt = 0.05
overwrite = False
stim_amp = 5000.0
mesh_unit = "cm"
dx = 0.05 * beat.units.ureg("cm").to(mesh_unit).magnitude
L = 1.0 * beat.units.ureg("cm").to(mesh_unit).magnitude
mesh = beat.geometry.get_3D_slab_mesh(Lx=L, Ly=dx, Lz=dx, dx=dx / 5, dim=dimension)
ffun = dolfin.MeshFunction("size_t", mesh, mesh.topology().dim() - 1)
ffun.set_all(0)
stim_domain = dolfin.CompiledSubDomain("x[0] <= DOLFIN_EPS")
marker = 1
stim_domain.mark(ffun, marker)
cfun = dolfin.MeshFunction("size_t", mesh, mesh.topology().dim())
cfun.set_all(0)
endo = dolfin.CompiledSubDomain("x[0] < L / 3", L=L)
epi = dolfin.CompiledSubDomain("x[0] > 2 * L / 3", L=L)
endo.mark(cfun, 1)
epi.mark(cfun, 2)
V = dolfin.FunctionSpace(mesh, "CG", 1)
pyvista.start_xvfb()
plotter_markers = pyvista.Plotter()
topology, cell_types, x = beat.viz.create_vtk_structures(V)
grid = pyvista.UnstructuredGrid(topology, cell_types, x)
grid["markers"] = cfun.array()
plotter_markers.add_mesh(grid, show_edges=True)
if mesh.geometric_dimension() == 2:
plotter_markers.view_xy()
/usr/lib/python3/dist-packages/pyvista/plotting/utilities/xvfb.py:48: PyVistaDeprecationWarning: This function is deprecated and will be removed in future version of PyVista. Use vtk-osmesa instead.
warnings.warn(
if not pyvista.OFF_SCREEN:
plotter_markers.show()
else:
figure_as_array = plotter_markers.screenshot("markers.png")
Interpolate meshfunction to a CG 1 function
cfun_DG = dolfin.Function(dolfin.FunctionSpace(mesh, "DG", 0))
cfun_DG.vector()[:] = cfun.array()
cfun_func = dolfin.Function(V)
cfun_func.interpolate(cfun_DG)
Calling FFC just-in-time (JIT) compiler, this may take some time.
g_il = 0.16069
g_el = 0.625
g_it = 0.04258
g_et = 0.236
f0, s0, n0 = beat.geometry.get_3D_slab_microstructure(transverse)
markers = {"ENDO": (marker, 2)}
data = Geometry(
mesh=mesh,
ffun=ffun,
markers=markers,
f0=f0,
s0=s0,
n0=n0,
)
save_freq = round(save_every_ms / dt)
print("Running model")
# Load the model
model_path = Path("ORdmm_Land.py")
if not model_path.is_file():
print("Generate code for cell model")
here = Path.cwd()
ode = gotranx.load_ode(here / ".." / "odes" / "ORdmm_Land.ode")
code = gotranx.cli.gotran2py.get_code(
ode, scheme=[gotranx.schemes.Scheme.forward_generalized_rush_larsen]
)
model_path.write_text(code)
Running model
import ORdmm_Land
model = ORdmm_Land.__dict__
Surface to volume ratio
chi = 1400.0 * beat.units.ureg("cm**-1")
Membrane capacitance
C_m = 1.0 * beat.units.ureg("uF/cm**2")
print("Get steady states")
nbeats = 2 # Should be set to at least 200
init_states = {
0: beat.single_cell.get_steady_state(
fun=model["forward_generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=2),
outdir=results_folder / "mid",
BCL=1000,
nbeats=nbeats,
track_indices=[model["state_index"]("v"), model["state_index"]("cai")],
dt=0.05,
),
1: beat.single_cell.get_steady_state(
fun=model["forward_generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=0),
outdir=results_folder / "endo",
BCL=1000,
nbeats=nbeats,
track_indices=[
model["state_index"]("v"),
model["state_index"]("cai"),
model["state_index"]("nai"),
],
dt=0.05,
),
2: beat.single_cell.get_steady_state(
fun=model["forward_generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=1),
outdir=results_folder / "epi",
BCL=1000,
nbeats=nbeats,
track_indices=[model["state_index"]("v"), model["state_index"]("cai")],
dt=0.05,
),
}
# endo = 0, epi = 1, M = 2
parameters = {
0: model["init_parameter_values"](amp=0.0, celltype=2),
1: model["init_parameter_values"](amp=0.0, celltype=0),
2: model["init_parameter_values"](amp=0.0, celltype=1),
}
fun = {
0: model["forward_generalized_rush_larsen"],
1: model["forward_generalized_rush_larsen"],
2: model["forward_generalized_rush_larsen"],
}
v_index = {
0: model["state_index"]("v"),
1: model["state_index"]("v"),
2: model["state_index"]("v"),
}
Get steady states
time = dolfin.Constant(0.0)
I_s = beat.stimulation.define_stimulus(
mesh=data.mesh,
chi=chi,
time=time,
subdomain_data=data.ffun,
marker=markers["ENDO"][0],
amplitude=stim_amp,
mesh_unit=mesh_unit,
)
M = beat.conductivities.define_conductivity_tensor(
chi,
f0=data.f0,
g_il=g_il,
g_it=g_it,
g_el=g_el,
g_et=g_et,
)
Cannot determine geometric dimension from expression.
params = {"preconditioner": "sor", "use_custom_preconditioner": False}
pde = beat.MonodomainModel(
time=time,
mesh=data.mesh,
M=M,
I_s=I_s,
params=params,
C_m=C_m.to(f"uF/{mesh_unit}**2").magnitude,
)
Calling FFC just-in-time (JIT) compiler, this may take some time.
V_ode = dolfin.FunctionSpace(data.mesh, "Lagrange", 1)
ode = beat.odesolver.DolfinMultiODESolver(
v_ode=dolfin.Function(V_ode),
v_pde=pde.state,
markers=cfun_func,
num_states={i: len(s) for i, s in init_states.items()},
fun=fun,
init_states=init_states,
parameters=parameters,
v_index=v_index,
)
t = 0.0
solver = beat.MonodomainSplittingSolver(pde=pde, ode=ode)
fname = (results_folder / "V.xdmf").as_posix()
beat.postprocess.delete_old_file(fname)
def save(t):
v = solver.pde.state.vector().get_local()
print(f"Solve for {t=:.2f}, {v.max() =}, {v.min() = }")
with dolfin.XDMFFile(data.mesh.mpi_comm(), fname) as xdmf:
xdmf.parameters["functions_share_mesh"] = True
xdmf.parameters["rewrite_function_mesh"] = False
xdmf.write_checkpoint(
solver.pde.state,
"V",
float(t),
dolfin.XDMFFile.Encoding.HDF5,
True,
)
i = 0
while t < end_time + 1e-12:
# Make sure to save at the same time steps that is used by Ambit
if i % save_freq == 0:
save(t)
solver.step((t, t + dt))
i += 1
t += dt
Solve for t=0.00, v.max() =-87.89546959710835, v.min() = -88.02335419809276
Calling FFC just-in-time (JIT) compiler, this may take some time.
Solve for t=1.00, v.max() =90.16409861439406, v.min() = -88.02607403992604
Solve for t=2.00, v.max() =155.60126904958025, v.min() = -88.01650823270752
Solve for t=3.00, v.max() =68.36671573543047, v.min() = -87.99116982978647
Solve for t=4.00, v.max() =53.08328895560886, v.min() = -87.98948415995054
Solve for t=5.00, v.max() =46.05473148859784, v.min() = -87.98947969733771
Solve for t=6.00, v.max() =42.33592214942757, v.min() = -87.98946803594322
Solve for t=7.00, v.max() =40.27288970581523, v.min() = -87.9894506946784
Solve for t=8.00, v.max() =39.11429579662869, v.min() = -87.98943065512952
Solve for t=9.00, v.max() =38.46934749672168, v.min() = -87.98941065811567
Solve for t=10.00, v.max() =38.12392397401019, v.min() = -87.98939253207834
Solve for t=11.00, v.max() =37.95629764339345, v.min() = -87.98937719432936
Solve for t=12.00, v.max() =37.894936958331584, v.min() = -87.98936489735465
Solve for t=13.00, v.max() =37.89684555708988, v.min() = -87.98935549073683
Solve for t=14.00, v.max() =37.93587267716543, v.min() = -87.98934859787136
Solve for t=15.00, v.max() =37.99594075122863, v.min() = -87.98934345276115
Solve for t=16.00, v.max() =38.0685269141949, v.min() = -87.98933547815112
Solve for t=17.00, v.max() =38.14940670811743, v.min() = -87.98927443813628
Solve for t=18.00, v.max() =38.236910316056424, v.min() = -87.98856155635102
Solve for t=19.00, v.max() =38.32882485166466, v.min() = -87.9800083230519
Solve for t=20.00, v.max() =38.420776179119585, v.min() = -87.87654876901343
Compute conduction velocity#
threshold = 0.0
x0 = L * 0.25
x1 = L * 0.75
if mesh.geometry().dim() == 2:
p1 = (x0, dx * 0.5)
p2 = (x1, dx * 0.5)
else:
p1 = (x0, dx * 0.5, dx * 0.5) # type: ignore
p2 = (x1, dx * 0.5, dx * 0.5) # type: ignore
V = dolfin.FunctionSpace(mesh, "CG", 1)
v = dolfin.Function(V)
plotter_voltage = pyvista.Plotter()
grid = pyvista.UnstructuredGrid(topology, cell_types, x)
grid.point_data["V"] = v.vector().get_local()
viridis = plt.get_cmap("viridis")
sargs = dict(
title_font_size=25,
label_font_size=20,
fmt="%.2e",
color="black",
position_x=0.1,
position_y=0.8,
width=0.8,
height=0.1,
)
plotter_voltage.add_mesh(
grid,
show_edges=True,
lighting=False,
cmap=viridis,
scalar_bar_args=sargs,
clim=[-90.0, 40.0],
)
fname = (results_folder / "V.xdmf").as_posix()
times = beat.postprocess.load_timesteps_from_xdmf(fname)
t1 = np.inf
t2 = np.inf
gif_file = Path("voltage_slab_time.gif")
gif_file.unlink(missing_ok=True)
plotter_voltage.open_gif(gif_file.as_posix())
for i, t in times.items():
with dolfin.XDMFFile(mesh.mpi_comm(), fname) as xdmf:
xdmf.read_checkpoint(v, "V", i)
print(f"Read {t=:.2f}, {v(p1) =}, {v(p2) = }")
grid.point_data["V"] = v.vector().get_local()
plotter_voltage.write_frame()
if v(p1) > threshold:
t1 = min(t, t1)
if v(p2) > threshold:
t2 = min(t, t2)
plotter_voltage.close()
Read t=0.00, v(p1) =-88.0233541980927, v(p2) = -87.98947492929054
Read t=1.00, v(p1) =-88.01916535919312, v(p2) = -87.98777385785289
Read t=2.00, v(p1) =-88.0122932758793, v(p2) = -87.98437760692872
Read t=3.00, v(p1) =-87.94809486293272, v(p2) = -87.98235387336061
Read t=4.00, v(p1) =-86.96609578867819, v(p2) = -87.98122162805575
Read t=5.00, v(p1) =-75.96569557749254, v(p2) = -87.98057085330106
Read t=6.00, v(p1) =-0.70590631458738, v(p2) = -87.98018525103916
Read t=7.00, v(p1) =16.386604223802436, v(p2) = -87.97995121999216
Read t=8.00, v(p1) =22.617822639049997, v(p2) = -87.97980677979197
Read t=9.00, v(p1) =26.629830401140232, v(p2) = -87.9797166074543
Read t=10.00, v(p1) =29.481002056075567, v(p2) = -87.97965854657642
Read t=11.00, v(p1) =31.6529186307407, v(p2) = -87.97960319178219
Read t=12.00, v(p1) =33.318574068479194, v(p2) = -87.97934604829611
Read t=13.00, v(p1) =34.58179237520088, v(p2) = -87.9765700572088
Read t=14.00, v(p1) =35.529572438773286, v(p2) = -87.94403524105732
Read t=15.00, v(p1) =36.238234418341925, v(p2) = -87.56293397443041
Read t=16.00, v(p1) =36.77285919886456, v(p2) = -83.23439701297086
Read t=17.00, v(p1) =37.18546120049233, v(p2) = -40.49428275226854
Read t=18.00, v(p1) =37.514434784549124, v(p2) = 13.014507025791161
Read t=19.00, v(p1) =37.786126523385505, v(p2) = 20.353785419781815
Read t=20.00, v(p1) =38.01758927358179, v(p2) = 24.51184126691342
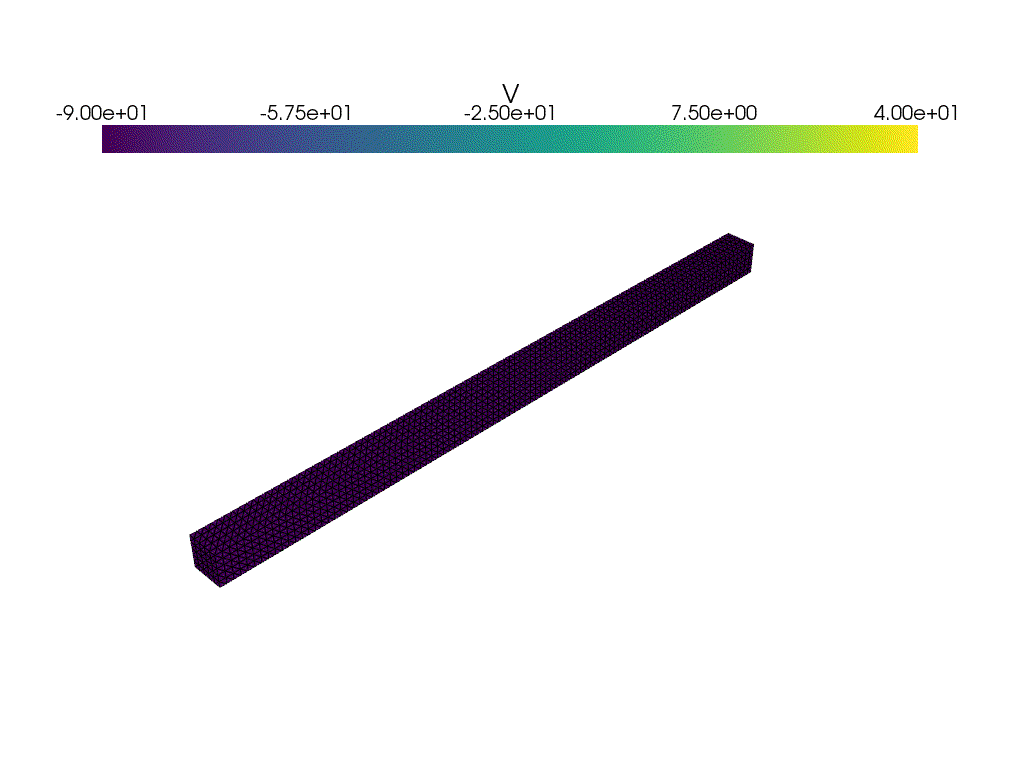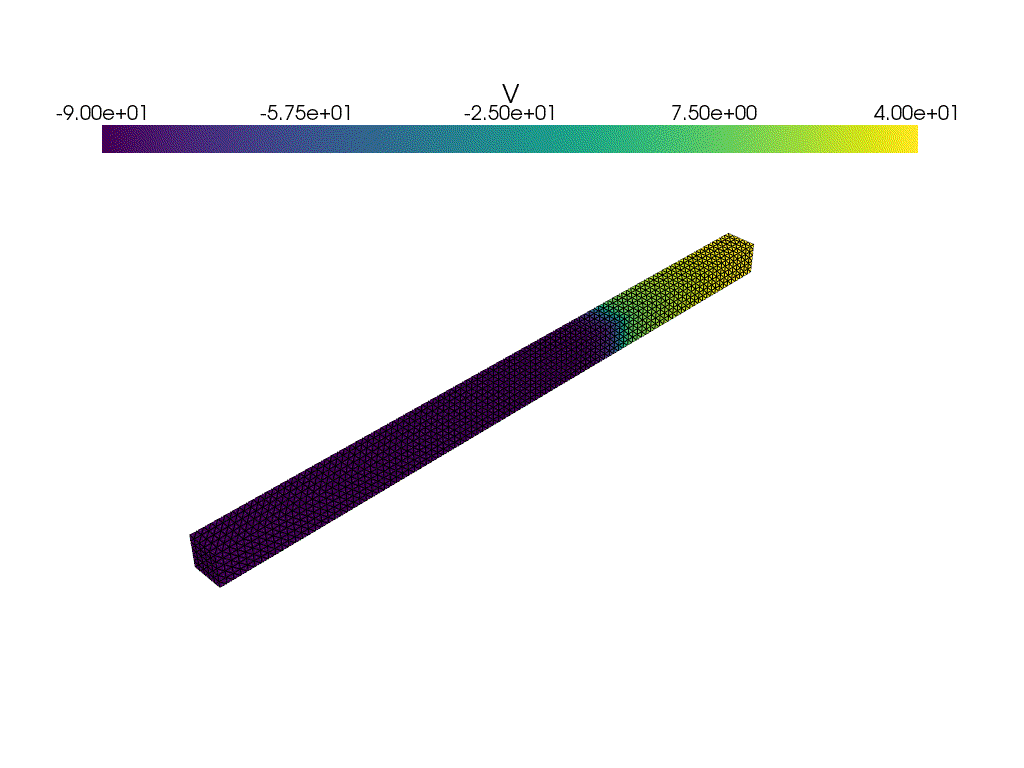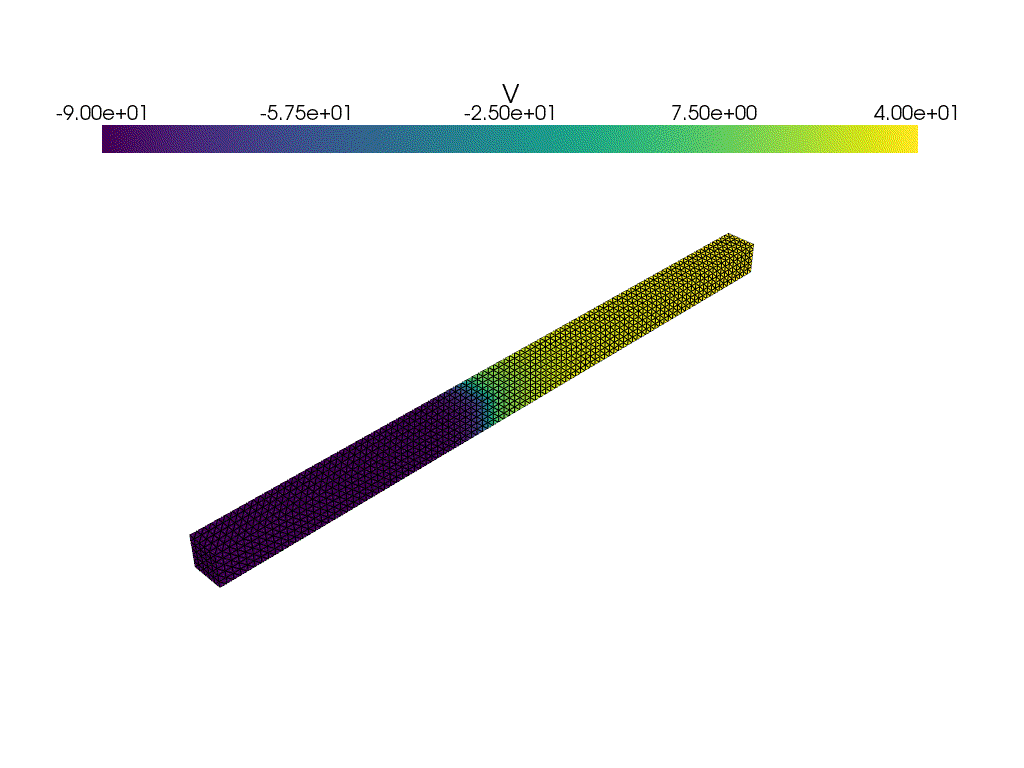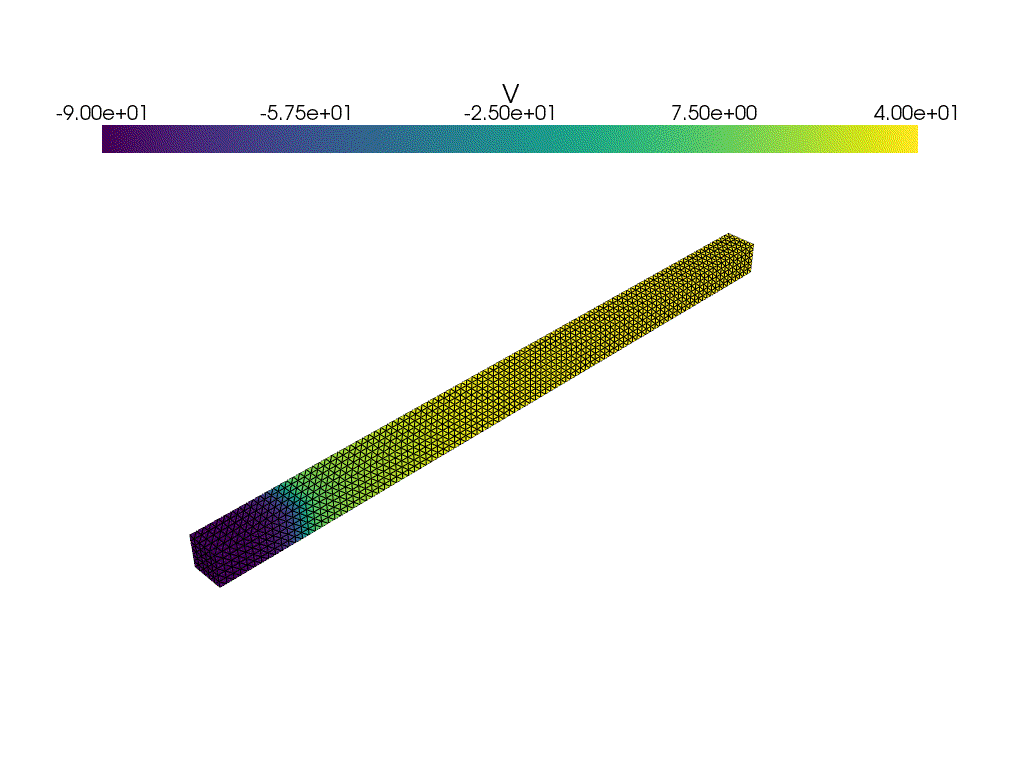
if not np.isclose(t1, t2):
cv = (x1 - x0) / (t2 - t1) * beat.units.ureg(f"{mesh_unit}/ms")
msg = (
f"Conduction velocity = {cv.magnitude:.3f} mm/ms or " #
f" {cv.to('m/s').magnitude:.3f} m/s or " #
f" {cv.to('cm/s').magnitude:.3f} cm/s" #
)
print(msg)
Conduction velocity = 0.045 mm/ms or 0.455 m/s or 45.455 cm/s
x0 = L * 2.0
if mesh.geometry().dim() == 2:
p = (x0, dx * 0.5)
else:
p = (x0, dx * 0.5, dx * 0.5) # type: ignore
V = dolfin.FunctionSpace(mesh, "CG", 1)
v = dolfin.Function(V)
phie = []
for i, t in times.items():
with dolfin.XDMFFile(mesh.mpi_comm(), fname) as xdmf:
xdmf.read_checkpoint(v, "V", i)
print(f"Read {t=:.2f}")
phie.append(beat.ecg.ecg_recovery(v=v, mesh=mesh, sigma_b=1.0, point=p))
Read t=0.00
Read t=1.00
Read t=2.00
Read t=3.00
Read t=4.00
Read t=5.00
Read t=6.00
Read t=7.00
Read t=8.00
Read t=9.00
Read t=10.00
Read t=11.00
Read t=12.00
Read t=13.00
Read t=14.00
Read t=15.00
Read t=16.00
Read t=17.00
Read t=18.00
Read t=19.00
Read t=20.00
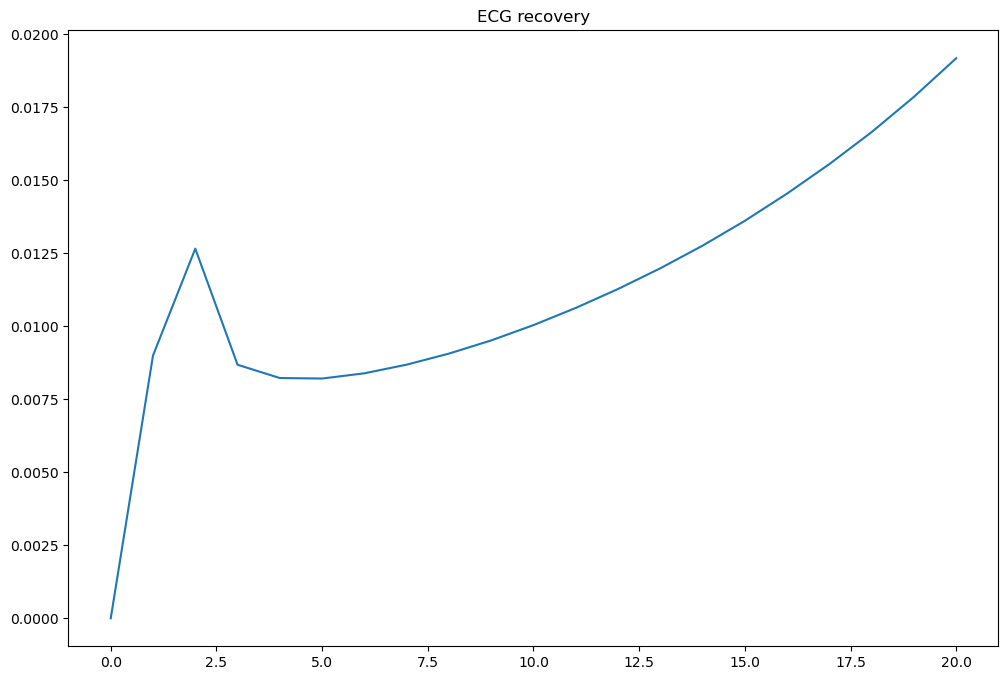
fig, ax = plt.subplots(1, 1, figsize=(12, 8))
ax.plot(times.values(), phie)
ax.set_title("ECG recovery")
fig.savefig(results_folder / "ecg_recovery.png")