Compiling a C-extension#
In this demo we will show how to set up a system that takes your .ode file, generates C-code, compiles the code just-in-time and imports the functions into python again. Note that there are several steps involved in this process we therefore split the code across two different modules utils.py and cmodel.py
To start with we will just run through an example how how this can be used.
First we import the modules utils where we add all the functionality as well as matplotlib for plotting
Note
The full source code for the all the files need for this demo (including utils.py) is found at the bottom of this document
import matplotlib.pyplot as plt
import utils
For this tutorial we will use a rather large system of ODE which simulated the electromechanics in cardiac cells that are based on the O’Hara-Rudy model for electrophysiology and the Land model. You can download the model in .ode format here
Next we load the model. This is function contains the functionality for generating code and compiling the C-extension. Currently it will also regenerate the code as well as recompiling the code code every time you run the code. It is also possible to only do this if the relevant files do not exist.
model = utils.load_model("ORdmm_Land.ode", rebuild=True, regenerate=True)
2026-03-04 06:22:43 [info ] Load ode ORdmm_Land.ode
2026-03-04 06:22:43 [info ] Num states 48
2026-03-04 06:22:43 [info ] Num parameters 139
CMake Deprecation Warning at CMakeLists.txt:1 (cmake_minimum_required):
Compatibility with CMake < 3.10 will be removed from a future version of
CMake.
Update the VERSION argument <min> value. Or, use the <min>...<max> syntax
to tell CMake that the project requires at least <min> but has been updated
to work with policies introduced by <max> or earlier.
-- The C compiler identification is GNU 11.4.0
-- Detecting C compiler ABI info
-- Detecting C compiler ABI info - done
-- Check for working C compiler: /usr/bin/cc - skipped
-- Detecting C compile features
-- Detecting C compile features - done
-- Configuring done (1.4s)
-- Generating done (0.0s)
-- Build files have been written to: /home/runner/work/gotranx/gotranx/examples/compile-c-extension/build_ORdmm_Land
[ 50%] Building C object CMakeFiles/ORdmm_Land.dir/demo.c.o
[100%] Linking C shared library lib/libORdmm_Land.so
[100%] Built target ORdmm_Land
2026-03-04 06:22:49 [info ] Load ode ORdmm_Land.ode
2026-03-04 06:22:49 [info ] Num states 48
2026-03-04 06:22:49 [info ] Num parameters 139
Next we get the initial states and parameters. The parameters we get back from initial_parameter_values are a numpy array. We also convert the parameters to a dictionary which is easier to work with since we can use the name of the parameter rather than the index.
y = model.initial_state_values()
# Get initial parameter values
p = model.initial_parameter_values()
parameters = model.parameter_values_to_dict(p)
Next we solve the model for 1000.0 milliseconds with a time step of 0.01 ms. Note that it would also be possible to make this loop in python (similar to the python API demo), however we will get a lot of performance gain if we instead do this loop in C.
# Simulate the model
sol = model.solve(0, 1000, dt=0.01, u0=y, parameters=parameters)
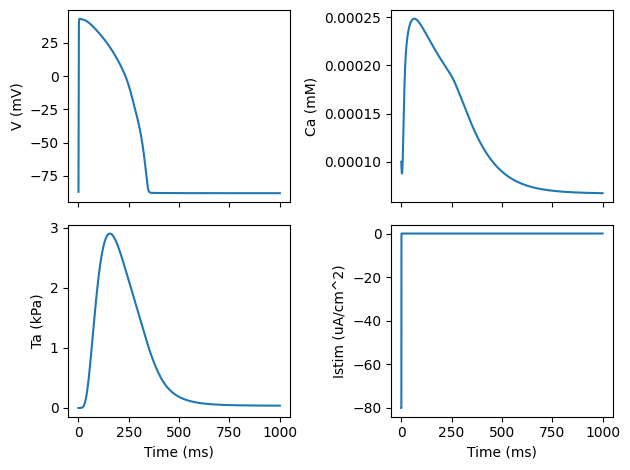
Now let us extract the state variables for the voltage and intracellular calcium
V = sol["v"]
Ca = sol["cai"]
as well as some monitor values
m = sol.monitor(["Ta", "Istim"])
Ta = m[:, 0]
Istim = m[:, 1]
and finally we plot the results
# Plot the results
fig, ax = plt.subplots(2, 2, sharex=True)
ax[0, 0].plot(sol.time, V)
ax[1, 0].plot(sol.time, Ta)
ax[0, 1].plot(sol.time, Ca)
ax[1, 1].plot(sol.time, Istim)
ax[1, 0].set_xlabel("Time (ms)")
ax[1, 1].set_xlabel("Time (ms)")
ax[0, 0].set_ylabel("V (mV)")
ax[1, 0].set_ylabel("Ta (kPa)")
ax[0, 1].set_ylabel("Ca (mM)")
ax[1, 1].set_ylabel("Istim (uA/cm^2)")
fig.tight_layout()
plt.show()
Source code#
utils.py#
import shutil
import subprocess as sp
from pathlib import Path
import numpy as np
from cmodel import CModel
import gotranx
HERE = Path(__file__).absolute().parent
MODEL_C_DIR = HERE
def cpath(odefile):
return (MODEL_C_DIR / str(odefile)).with_suffix(".h")
def cbuild_dir(model):
cfile = cpath(model)
return MODEL_C_DIR.joinpath(f"build_{cfile.stem}")
def load_model(ode_file, rebuild=True, regenerate=False):
# Check if ode_file is present
cfile = cpath(ode_file)
if not cfile.is_file() or regenerate:
gotran2c(ode_file)
if not cfile.is_file() or rebuild:
build_c(ode_file)
build_dir = cbuild_dir(ode_file)
lib = np.ctypeslib.load_library(next(build_dir.joinpath("lib").iterdir()), HERE)
ode = gotranx.load_ode(str(ode_file))
return CModel(lib, ode)
def build_c(model):
cfile = cpath(model)
with open(MODEL_C_DIR.joinpath("template.c"), "r") as f:
template = f.read()
include_str = f'#include "{cfile.name}"\n'
with open(MODEL_C_DIR.joinpath("demo.c"), "w") as f:
f.write(include_str + template)
model_name = cfile.stem
build_dir = cbuild_dir(model)
if build_dir.exists():
shutil.rmtree(build_dir)
build_dir.mkdir()
sp.check_call(["cmake", "-S", ".", "-B", str(build_dir), f"-DCELL_LIBFILE={model_name}"])
sp.check_call(["cmake", "--build", str(build_dir)])
def gotran2c(odefile):
ode = gotranx.load_ode(odefile)
# Generate code and generalized rush larsen scheme
code = gotranx.cli.gotran2c.get_code(
ode,
scheme=[
gotranx.schemes.Scheme.forward_generalized_rush_larsen,
gotranx.schemes.Scheme.forward_explicit_euler,
],
)
fname = Path(odefile).with_suffix(".h").name
(MODEL_C_DIR / fname).write_text(code)
cmodel.py#
from ctypes import c_char_p
from ctypes import c_double
from ctypes import c_int
from typing import Dict
from typing import List
from typing import NamedTuple
from typing import Optional
import numpy as np
class CModel:
def __init__(self, lib, ode):
self.lib = lib
# Get number of states and parameters from the C library
self.num_states = self.lib.state_count()
self.num_parameters = self.lib.parameter_count()
self.num_monitored = self.lib.monitor_count()
self._init_lib()
self.ode = ode
def __repr__(self) -> str:
return f"CModel({self.lib}, {self.ode})"
def __str__(self) -> str:
return (
f"Model named {self.ode.name} with {self.num_states} states, "
f"{self.num_parameters} parameters and {self.num_monitored} "
"monitored values"
)
def parameter_values_to_dict(
self,
parameter_values: np.ndarray,
) -> Dict[str, float]:
"""Convert the parameter values using the ordered from the C library
to a dictionary with parameter names as keys and the values as values"""
names = self.parameter_names
values = [parameter_values[self.parameter_index(name)] for name in names]
return dict(zip(names, values))
def state_values_to_dict(self, state_values: np.ndarray) -> Dict[str, float]:
"""Convert the state values using the ordered from the C library
to a dictionary with state names as keys and the values as values"""
names = self.state_names
values = [state_values[self.state_index(name)] for name in names]
return dict(zip(names, values))
def parameter_dict_to_array(self, parameter_dict: Dict[str, float]) -> np.ndarray:
"""Convert the a dictionary of parameters to an array of values
with the correct order.
"""
values = self.initial_parameter_values()
for name, value in parameter_dict.items():
values[self.parameter_index(name)] = value
return values
def state_dict_to_array(self, state_dict: Dict[str, float]) -> np.ndarray:
"""Convert the a dictionary of states to an array of values
with the correct order.
"""
values = self.initial_state_values()
for name, value in state_dict.items():
values[self.state_index(name)] = value
return values
def default_parameters(self) -> Dict[str, float]:
"""Return the default parameter as a dictionary where the
keys are the parameter names and the values"""
return self.parameter_values_to_dict(self.initial_parameter_values())
def default_initial_states(self) -> Dict[str, float]:
"""Return the default initial as a dictionary where the
keys are the parameter names and the values"""
return self.state_values_to_dict(self.initial_state_values())
@property
def parameter_names(self) -> List[str]:
"""List of parameters names"""
return [p.name for p in self.ode.parameters]
@property
def state_names(self) -> List[str]:
"""List of state names"""
return [p.name for p in self.ode.states]
@property
def monitor_names(self) -> List[str]:
"""List of monitor names"""
return [expr.name for expr in self.ode.intermediates + self.ode.state_derivatives]
def _init_lib(self):
"""
Make sure that arrays passed to C is of the correct types.
"""
float64_array = np.ctypeslib.ndpointer(
dtype=c_double,
ndim=1,
flags="contiguous",
)
int32_array = np.ctypeslib.ndpointer(
dtype=c_int,
ndim=1,
flags="contiguous",
)
float64_array_2d = np.ctypeslib.ndpointer(
dtype=c_double,
ndim=2,
flags="contiguous",
)
self.lib.init_state_values.restype = None # void
self.lib.init_state_values.argtypes = [float64_array]
self.lib.init_parameter_values.restype = None # void
self.lib.init_parameter_values.argtypes = [float64_array]
self.lib.state_index.restype = c_int
self.lib.state_index.argtypes = [c_char_p] # state_name
self.lib.parameter_index.restype = c_int
self.lib.parameter_index.argtypes = [c_char_p] # state_name
self.lib.monitor_index.restype = c_int
self.lib.monitor_index.argtypes = [c_char_p] # state_name
self.lib.monitored_values.restype = None
self.lib.monitored_values.argtypes = [
float64_array_2d, # monitored
float64_array_2d, # states
float64_array, # parameters
float64_array, # u
float64_array, # t_values
c_int, # num_timesteps
int32_array, # indices
c_int, # num_indices
]
advance_functions = [
self.lib.forward_explicit_euler,
self.lib.forward_generalized_rush_larsen,
]
for func in advance_functions:
func.restype = None # void
func.argtypes = [
float64_array, # u
c_double, # t
c_double, # dt
float64_array, # parameters
]
solve_functions = [
self.lib.ode_solve_forward_euler,
self.lib.ode_solve_rush_larsen,
]
for func in solve_functions:
func.restype = None # void
func.argtypes = [
float64_array, # u
float64_array, # parameters
float64_array_2d, # u_values
float64_array, # t_values
c_int, # num_timesteps
c_double, # dt
]
def advance_ODEs(
self,
states: np.ndarray,
t: float,
dt: float,
parameters: np.ndarray,
scheme="GRL1",
) -> np.ndarray:
u = states.copy()
if scheme == "GRL1":
self.lib.forward_generalized_rush_larsen(u, t, dt, parameters)
elif scheme == "FE":
self.lib.forward_explicit_euler(u, t, dt, parameters)
else:
raise ValueError(f"Unknown scheme {scheme}")
return u
def monitor(
self,
names: list[str],
states: np.ndarray,
t: np.ndarray,
parameters: Optional[Dict[str, float]] = None,
) -> np.ndarray:
"""Return a single monitored value
Parameters
----------
names : list[str]
Names of monitored values
states : np.ndarray
The states
t : np.ndarray
The time steps
parameters : Dict[str, float], optional
Dictionary with initial parameters, by default None
Returns
-------
np.ndarray
The values of the monitors
"""
indices = np.array([self.monitor_index(name) for name in names], dtype=np.int32)
parameter_values = self._get_parameter_values(parameters=parameters)
u = np.zeros(self.num_states, dtype=np.float64)
monitored_values = np.zeros((t.size, len(names)), dtype=np.float64)
self.lib.monitored_values(
monitored_values,
states,
parameter_values,
u,
t,
t.size,
indices,
len(indices),
)
return monitored_values
def state_index(self, state_name: str) -> int:
"""Given a name of a state, return the index of it.
Arguments
---------
state_name : str
Name of the state
Returns
-------
int
The index of the given state
Note
----
To list all possible states see `BaseModel.state_names`
"""
assert isinstance(state_name, str)
if state_name not in self.state_names:
raise ValueError(f"Invalid state name {state_name!r}")
state_name_bytestring = state_name.encode()
return self.lib.state_index(state_name_bytestring)
def parameter_index(self, parameter_name: str) -> int:
"""Given a name of a parameter, return the index of it.
Arguments
---------
parameter_name : str
Name of the parameter
Returns
-------
int
The index of the given parameter
"""
assert isinstance(parameter_name, str)
if parameter_name not in self.parameter_names:
raise ValueError(f"Invalid parameter name {parameter_name!r}")
parameter_name_bytestring = parameter_name.encode()
return self.lib.parameter_index(parameter_name_bytestring)
def monitor_index(self, monitor_name: str) -> int:
"""Given a name of a monitored expression, return the index of it.
Arguments
---------
monitor_name : str
Name of the monitored expression
Returns
-------
int
The index of the given monitored expression
"""
assert isinstance(monitor_name, str)
if monitor_name not in self.monitor_names:
raise ValueError(f"Invalid monitor name {monitor_name!r}")
monitor_name_bytestring = monitor_name.encode()
return self.lib.monitor_index(monitor_name_bytestring)
def initial_parameter_values(self) -> np.ndarray:
"""Return the default parameters as a numpy array"""
parameters = np.zeros(self.num_parameters, dtype=np.float64)
self.lib.init_parameter_values(parameters)
return parameters
def initial_state_values(self, **values) -> np.ndarray:
"""Return the default initial states as a numpy array"""
states = np.zeros(self.num_states, dtype=np.float64)
self.lib.init_state_values(states)
for key, value in values.items():
states[self.state_index(key)] = value
return states
def _get_parameter_values(
self,
parameters: Optional[Dict[str, float]],
verbose: bool = False,
) -> np.ndarray:
parameter_values = self.initial_parameter_values()
if parameters is not None:
assert isinstance(parameters, dict)
for name, new_value in parameters.items():
index = self.parameter_index(name)
old_value = parameter_values[index]
if old_value != new_value:
parameter_values[index] = new_value
if verbose:
print(
f"Update parameter {name} from " f"{old_value} to {new_value}",
)
return parameter_values
def solve(
self,
t_start: float,
t_end: float,
dt: float,
num_steps: Optional[int] = None,
method: str = "GRL1",
u0: Optional[np.ndarray] = None,
parameters: Optional[Dict[str, float]] = None,
verbose: bool = False,
):
"""Solve the model
Parameters
----------
t_start : float
Time at start point
t_end : float
Time at end point
dt : float
Time step for solver
num_steps : Optional[int], optional
Number of steps to use, by default None
method : str, optional
Scheme for solving the ODE. Options are
'GRL1' (first order generalized Rush Larsen) or
'FE' (forward euler), by default "GRL1"
u0 : Optional[np.ndarray], optional
Initial state variables. If none is provided then
the default states will be used, by default None.
parameters : Optional[Dict[str, float]], optional
Parameter for the model. If none is provided then
the default parameters will be used, by default None.
verbose : bool, optional
Print more output, by default False
"""
parameter_values = self._get_parameter_values(
parameters=parameters,
verbose=verbose,
)
if not isinstance(dt, float):
dt = float(dt)
if num_steps is not None:
assert isinstance(num_steps, int)
t_end = dt * num_steps
else:
num_steps = round((t_end - t_start) / dt)
t_values = np.linspace(t_start, t_end, num_steps + 1)
if u0 is not None:
assert len(u0) == self.num_states
else:
u0 = np.zeros(self.num_states, dtype=np.float64)
self.lib.init_state_values(u0)
u_values = np.zeros((num_steps + 1, u0.shape[0]), dtype=np.float64)
u_values[0, :] = u0[:]
if method == "FE":
self.lib.ode_solve_forward_euler(
u0,
parameter_values,
u_values,
t_values,
num_steps,
dt,
)
elif method == "GRL1":
self.lib.ode_solve_rush_larsen(
u0,
parameter_values,
u_values,
t_values,
num_steps,
dt,
)
else:
raise ValueError(f"Invalid method {method}")
return Solution(
time=t_values,
u=u_values,
parameter_values=parameter_values,
model=self,
)
class Solution(NamedTuple):
time: np.ndarray
u: np.ndarray
parameter_values: np.ndarray
model: CModel
@property
def parameters(self):
return self.model.parameter_values_to_dict(self.parameter_values)
def keys(self):
return self.model.state_names
def monitor_keys(self):
return self.model.monitor_names
def monitor(self, names: list[str]):
return self.model.monitor(
names,
self.u,
self.time,
parameters=self.parameters,
)
def __getitem__(self, name):
if name not in self.keys():
raise KeyError("Key {name} not a valid state name")
index = self.model.state_index(name)
return self.u[:, index]
template.c#
#include <math.h>
#include <stdio.h>
#include <stdlib.h>
#include <string.h>
#include <time.h>
void ode_solve_forward_euler(double* u, const double* parameters,
double* u_values, double* t_values,
int num_timesteps, double dt) {
double t;
int save_it = 1;
int it, j;
double u_temp[NUM_STATES];
for (it = 1; it <= num_timesteps; it++) {
t = t_values[it - 1];
forward_explicit_euler(u, t, dt, parameters, u_temp);
for (j = 0; j < NUM_STATES; j++) {
u_values[save_it * NUM_STATES + j] = u_temp[j];
u[j] = u_temp[j];
}
save_it++;
}
}
void ode_solve_rush_larsen(double* u, const double* parameters,
double* u_values, double* t_values,
int num_timesteps, double dt) {
double t;
int save_it = 1;
int it, j;
double u_temp[NUM_STATES];
for (it = 1; it <= num_timesteps; it++) {
t = t_values[it - 1];
forward_generalized_rush_larsen(u, t, dt, parameters, u_temp);
for (j = 0; j < NUM_STATES; j++) {
u_values[save_it * NUM_STATES + j] = u_temp[j];
u[j] = u_temp[j];
}
save_it++;
}
}
void monitored_values(double* monitored, double* states,
double* parameters, double* u,
double* t_values, int length, int *indices, int num) {
double t;
int i, j;
double m_temp[NUM_MONITORED];
for (i = 0; i < length; i++) {
t = t_values[i];
for (j = 0; j < NUM_STATES; j++) {
u[j] = states[i * NUM_STATES + j];
}
monitor_values(t, u, parameters, m_temp);
for (j = 0; j < num; j++) {
monitored[i * num + j] = m_temp[indices[j]];
}
}
}
int state_count() {
return NUM_STATES;
}
int parameter_count() {
return NUM_PARAMS;
}
int monitor_count() {
return NUM_MONITORED;
}
int main(int argc, char* argv[]) {
double t_start = 0;
double dt = 0.02E-3;
int num_timesteps = (int)1000000;
if (argc > 1) {
num_timesteps = atoi(argv[1]);
printf("num_timesteps set to %d\n", num_timesteps);
if (num_timesteps <= 0) {
exit(EXIT_FAILURE);
}
}
unsigned int num_states = NUM_STATES;
size_t states_size = num_states * sizeof(double);
unsigned int num_parameters = NUM_PARAMS;
size_t parameters_size = num_parameters * sizeof(double);
double* states = malloc(states_size);
double* states_values = malloc(states_size);
double* parameters = malloc(parameters_size);
init_parameter_values(parameters);
double t = t_start;
struct timespec timestamp_start, timestamp_now;
double time_elapsed;
// forward euler
printf("Scheme: Forward Euler\n");
clock_gettime(CLOCK_MONOTONIC_RAW, ×tamp_start);
init_state_values(states);
int it;
for (it = 0; it < num_timesteps; it++) {
forward_explicit_euler(states, t, dt, parameters, states_values);
t += dt;
}
clock_gettime(CLOCK_MONOTONIC_RAW, ×tamp_now);
time_elapsed = timestamp_now.tv_sec - timestamp_start.tv_sec + 1E-9 * (timestamp_now.tv_nsec - timestamp_start.tv_nsec);
printf("Computed %d time steps in %g s. Time steps per second: %g\n",
num_timesteps, time_elapsed, num_timesteps / time_elapsed);
printf("\n");
// Rush Larsen
printf("Scheme: Rush Larsen (exp integrator on all gates)\n");
clock_gettime(CLOCK_MONOTONIC_RAW, ×tamp_start);
init_state_values(states);
for (it = 0; it < num_timesteps; it++) {
forward_generalized_rush_larsen(states, t, dt, parameters, states_values);
t += dt;
}
clock_gettime(CLOCK_MONOTONIC_RAW, ×tamp_now);
time_elapsed = timestamp_now.tv_sec - timestamp_start.tv_sec + 1E-9 * (timestamp_now.tv_nsec - timestamp_start.tv_nsec);
printf("Computed %d time steps in %g s. Time steps per second: %g\n",
num_timesteps, time_elapsed, num_timesteps / time_elapsed);
printf("\n");
free(states);
free(parameters);
return 0;
}
CMakeLists.txt#
cmake_minimum_required(VERSION 3.9 FATAL_ERROR)
project(cellmodel LANGUAGES C)
set(CMAKE_POSITION_INDEPENDENT_CODE True)
if("${PROJECT_BINARY_DIR}" STREQUAL "${PROJECT_SOURCE_DIR}")
message(FATAL_ERROR "You cannot build in the source directory. Please run cmake from a subdirectory called 'build'")
endif()
if(NOT CMAKE_BUILD_TYPE)
set(CMAKE_BUILD_TYPE Release)
endif()
set(CMAKE_C_FLAGS_RELEASE "-O3")
set(CMAKE_RUNTIME_OUTPUT_DIRECTORY ${PROJECT_BINARY_DIR}/bin)
set(CMAKE_LIBRARY_OUTPUT_DIRECTORY ${PROJECT_BINARY_DIR}/lib)
add_library(${CELL_LIBFILE} SHARED demo.c)
install(TARGETS ${CELL_LIBFILE} DESTINATION lib)