Pacing traing#
In this demo we will repeat the same setup as in the PVC demo but we will stimulate the cells on the left boundary of the cable and a rapid pace. First we do the necceary imports
Next we set the output directory for the results and define the geometry. Here we specify an interval mesh of 200 cells with 0.015 cm between each cell
here = Path.cwd()
outdir = here / "results-pacing-train"
mesh_unit = "cm"
dx = 0.015
num_cells = 200
L = num_cells * dx
comm = MPI.COMM_WORLD
mesh = dolfinx.mesh.create_interval(comm, num_cells, (0, L))
We will use the tensusscher panfilov model
model_path = Path("tentusscher_panfilov_2006_epi_cell.py")
if not model_path.is_file():
here = Path.cwd()
ode = gotranx.load_ode(
here
/ ".."
/ "odes"
/ "tentusscher_panfilov_2006"
/ "tentusscher_panfilov_2006_epi_cell.ode",
)
code = gotranx.cli.gotran2py.get_code(
ode,
scheme=[gotranx.schemes.Scheme.generalized_rush_larsen],
)
model_path.write_text(code)
import tentusscher_panfilov_2006_epi_cell
model = tentusscher_panfilov_2006_epi_cell.__dict__
Here we can also specify whether the stimulation should be on one side or if we should stimulate all cell at once. We can also specify an end time for the simulation.
# Change this to run the simulations for longer
end_time = 500.0
We specify a diffusion coefficient and a membrane capacitance
D = 0.0005 * beat.units.ureg("cm**2 / ms")
Cm = 1.0 * beat.units.ureg("uF/cm**2")
Next we run a single cell model with the to get the correct steady state solutions. We run this for 50 beats with a stimulation every 100.0 ms (so this is quite rapid)
stim_period = 100.0
parameters = model["init_parameter_values"](stim_period=stim_period)
dt = 0.01
nbeats = 50
fun = model["generalized_rush_larsen"]
y = get_steady_state(
fun=fun,
init_states=model["init_state_values"](),
parameters=parameters,
outdir=outdir / "prebeats",
BCL=500,
nbeats=nbeats,
track_indices=[model["state_index"]("V"), model["state_index"]("Ca_i")],
dt=dt,
)
# -
We stimulate the left side of the cable stimulate after 100 ms with period of 500 ms
stim_duration = 2.0
stim_amp = dolfinx.fem.Constant(mesh, 0.0)
time = dolfinx.fem.Constant(mesh, 0.0)
start = 10.0
# Turn off stimulation in the ODE model
parameters = model["init_parameter_values"](stim_amplitude=0.0)
def S1_subdomain(x):
return x[0] <= 2 * dx
facets = dolfinx.mesh.locate_entities_boundary(
mesh,
mesh.topology.dim - 1,
S1_subdomain,
)
marker = 1
subdomain_data = dolfinx.mesh.meshtags(
mesh,
mesh.topology.dim - 1,
facets,
np.full(len(facets), marker, dtype=np.int32),
)
ds = ufl.Measure("ds", domain=mesh, subdomain_data=subdomain_data)(marker)
I_s = beat.base_model.Stimulus(dZ=ds, expr=stim_amp, marker=marker)
Now we would like to change the conductances for the cells in the right most part of the cable. We choose a first order Lagrange space for this, so that we have one set of parameters for each cells, and then we copy over the default parameters
V_ode = dolfinx.fem.functionspace(mesh, ("Lagrange", 1))
parameters_ode = np.zeros(
(len(parameters), V_ode.dofmap.index_map.size_local),
dtype=np.float64,
)
parameters_ode.T[:] = parameters
Next we set the values og \(g_Kr\) to zero in the right part of the cable
g_Kr_index = model["parameter_index"]("g_Kr")
g_Kr_value = parameters[g_Kr_index]
g_Kr = dolfinx.fem.Function(V_ode)
g_Kr.interpolate(
dolfinx.fem.Expression(
ufl.conditional(ufl.ge(X[0], L / 2), 0.0, g_Kr_value),
V_ode.element.interpolation_points(),
),
)
parameters_ode[g_Kr_index, :] = g_Kr.x.array
and similar for the conductance \(g_Ks\)
g_Ks_index = model["parameter_index"]("g_Ks")
g_Ks_value = parameters[g_Ks_index]
g_Ks = dolfinx.fem.Function(V_ode)
g_Ks.interpolate(
dolfinx.fem.Expression(
ufl.conditional(ufl.ge(X[0], L / 2), 0.0, g_Ks_value),
V_ode.element.interpolation_points(),
),
)
parameters_ode[g_Ks_index, :] = g_Ks.x.array
Finally we set up the models
pde = beat.MonodomainModel(
time=time,
mesh=mesh,
M=D.magnitude,
I_s=I_s,
C_m=Cm.magnitude,
)
ode = beat.odesolver.DolfinODESolver(
v_ode=dolfinx.fem.Function(V_ode),
v_pde=pde.state,
fun=fun,
init_states=y,
parameters=parameters_ode,
num_states=len(y),
v_index=model["state_index"]("V"),
)
solver = beat.MonodomainSplittingSolver(pde=pde, ode=ode)
checkpointfname = outdir / "slab_checkpoint.bp"
shutil.rmtree(checkpointfname, ignore_errors=True)
adios4dolfinx.write_mesh(checkpointfname, mesh)
def save(t):
v = solver.pde.state.x.array
if t % 100.0 == 0:
print(f"Solve for {t=:.2f}, {v.max() =}, {v.min() =}")
adios4dolfinx.write_function(checkpointfname, solver.pde.state, time=t, name="v")
and solve it
t = 0.0
save_freq = int(1.0 / dt)
i = 0
done_stimulating = False
while t < end_time + 1e-12:
# Make sure to save at the same time steps that is used by Ambit
if t > start and (t - start) % stim_period < stim_duration:
stim_amp.value = 1.0
else:
stim_amp.value = 0.0
if i % save_freq == 0:
save(t)
if t > 1000 and not done_stimulating:
ode.parameters[model["parameter_index"]("stim_amplitude"), :] = 0.0
I_s.assign(0.0)
done_stimulating = True
solver.step((t, t + dt))
i += 1
t += dt
Solve for t=0.00, v.max() =np.float64(-84.13338382291985), v.min() =np.float64(-84.13338382291985)
def post_process(dx, outdir):
mesh = adios4dolfinx.read_mesh(comm=comm, filename=checkpointfname)
V = dolfinx.fem.functionspace(mesh, ("P", 1))
v = dolfinx.fem.Function(V)
times = beat.postprocess.read_timestamps(comm, checkpointfname, "v")
cellnr = [0, 25, 50, 75, 100, 125, 150, 175, 200]
cellnr = np.arange(0, 200, 10)
p1 = 25 * dx
p2 = 175 * dx
dp = 150 * dx * beat.units.ureg("cm")
tp1 = np.inf
tp2 = np.inf
points = cellnr * dx
p1p2 = [p1, p2]
if not (outdir / "traces.npy").is_file():
traces = np.zeros((len(times), len(cellnr)))
for i, ti in enumerate(times):
adios4dolfinx.read_function(checkpointfname, v, time=ti, name="v")
traces[i, :] = scifem.evaluate_function(
v,
np.expand_dims(points, 1),
).squeeze()
vp1p2 = scifem.evaluate_function(v, np.expand_dims(p1p2, 1)).squeeze()
if vp1p2[0] > 0.0 and tp1 == np.inf:
tp1 = ti
if vp1p2[0] > 0.0 and tp2 == np.inf:
tp2 = ti
np.save(outdir / "traces.npy", traces)
np.save(outdir / "times.npy", times)
(outdir / "cv.text").write_text(f"{tp1} {tp2}")
traces = np.load(outdir / "traces.npy")
t = np.load(outdir / "times.npy")
tp1, tp2 = np.loadtxt(outdir / "cv.text")
tp1 *= beat.units.ureg("ms")
tp2 *= beat.units.ureg("ms")
if not np.isclose(tp1, tp2):
cv = dp / (tp2 - tp1)
print(
f"Conduction velocity:: {cv.to('cm/ms').magnitude} cm/ms "
f"= {cv.to('m/s').magnitude} m/s",
)
# Plot 3D plot for all traces
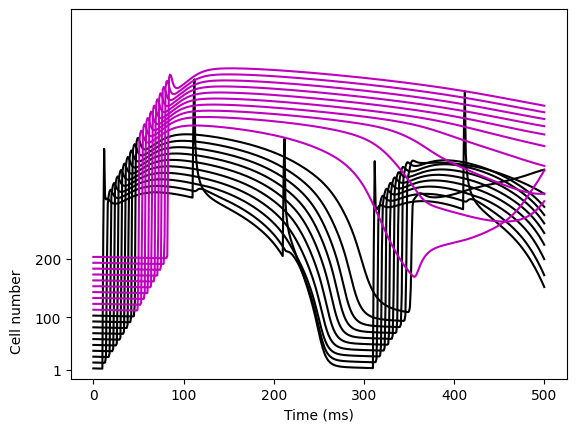
fig, ax = plt.subplots()
for i, cell_index in enumerate(cellnr):
color = "k" if cell_index < 100 else "m"
ax.plot(t, cell_index / 3 * np.ones_like(t) + traces[:, i], color=color)
ax.set_xlabel("Time (ms)")
ax.set_yticks([-22, -55, -85])
ax.set_yticklabels([200, 100, 1])
fig.text(x=0.03, y=0.17, s="Cell number", rotation=90)
ax.set_ylim(-90, 120)
fig.savefig(outdir / "V_3d.png")
post_process(dx, outdir)