Conduction velocity and ECG for slabs#
In this demo we will show how to compute conduction velocity and ECG for a Slab geometry.
import adios4dolfinx
import beat.postprocess
from beat.geometry import Geometry
comm = MPI.COMM_WORLD
results_folder = Path("results-slab")
results_folder.mkdir(exist_ok=True)
save_every_ms = 1.0
transverse = False
end_time = 20.0
dt = 0.05
overwrite = False
stim_amp = 5000.0
mesh_unit = "cm"
dx = 0.05 * beat.units.ureg("cm").to(mesh_unit).magnitude
L = 1.0 * beat.units.ureg("cm").to(mesh_unit).magnitude
mesh = beat.geometry.get_3D_slab_mesh(comm=comm, Lx=L, Ly=dx, Lz=dx, dx=dx / 5)
tol = 1.0e-8
marker = 1
def S1_subdomain(x):
return x[0] <= tol
facets = dolfinx.mesh.locate_entities_boundary(
mesh,
mesh.topology.dim - 1,
S1_subdomain,
)
ffun = dolfinx.mesh.meshtags(
mesh,
mesh.topology.dim - 1,
facets,
np.full(len(facets), marker, dtype=np.int32),
)
V = dolfinx.fem.functionspace(mesh, ("P", 1))
plotter_markers = pyvista.Plotter()
grid = pyvista.UnstructuredGrid(*dolfinx.plot.vtk_mesh(V))
plotter_markers.add_mesh(grid, show_edges=True)
if mesh.geometry.dim == 2:
plotter_markers.view_xy()
2026-03-04 06:42:19.998 ( 0.715s) [ 7F7DC331B140]vtkXOpenGLRenderWindow.:1460 WARN| bad X server connection. DISPLAY=
if not pyvista.OFF_SCREEN:
plotter_markers.show()
else:
plotter_markers.screenshot(results_folder / "markers.png")
cfun_func = dolfinx.fem.Function(V)
cfun_func.interpolate(endo_epi)
plotter_markers = pyvista.Plotter()
grid = pyvista.UnstructuredGrid(*dolfinx.plot.vtk_mesh(V))
grid.point_data["V"] = cfun_func.x.array
plotter_markers.add_mesh(grid, show_edges=True)
if mesh.geometry.dim == 2:
plotter_markers.view_xy()
if not pyvista.OFF_SCREEN:
plotter_markers.show()
else:
plotter_markers.screenshot(results_folder / "endo_epi.png")
with dolfinx.io.XDMFFile(comm, results_folder / "ffun.xdmf", "w") as xdmf:
xdmf.write_mesh(mesh)
xdmf.write_meshtags(ffun, mesh.geometry)
with dolfinx.io.XDMFFile(comm, results_folder / "endo_epi.xdmf", "w") as xdmf:
xdmf.write_mesh(mesh)
xdmf.write_function(cfun_func)
g_il = 0.16069
g_el = 0.625
g_it = 0.04258
g_et = 0.236
f0, s0, n0 = beat.geometry.get_3D_slab_microstructure(mesh, transverse)
markers = {"ENDO": (marker, 2)}
data = Geometry(
mesh=mesh,
ffun=ffun,
markers=markers,
f0=f0,
s0=s0,
n0=n0,
)
save_freq = round(save_every_ms / dt)
print("Running model")
# Load the model
model_path = Path("ToRORd_dynCl_endo.py")
if not model_path.is_file():
print("Generate code for cell model")
here = Path.cwd()
ode = gotranx.load_ode(here / ".." / "odes" / "torord" / "ToRORd_dynCl_endo.ode")
code = gotranx.cli.gotran2py.get_code(
ode,
scheme=[gotranx.schemes.Scheme.generalized_rush_larsen],
)
model_path.write_text(code)
Running model
import ToRORd_dynCl_endo
model = ToRORd_dynCl_endo.__dict__
Surface to volume ratio
chi = 1400.0 * beat.units.ureg("cm**-1")
Membrane capacitance
C_m = 1.0 * beat.units.ureg("uF/cm**2")
print("Get steady states")
nbeats = 2 # Should be set to at least 200
init_states = {
0: beat.single_cell.get_steady_state(
fun=model["generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=2),
outdir=results_folder / "mid",
BCL=1000,
nbeats=nbeats,
track_indices=[model["state_index"]("v"), model["state_index"]("cai")],
dt=0.05,
),
1: beat.single_cell.get_steady_state(
fun=model["generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=0),
outdir=results_folder / "endo",
BCL=1000,
nbeats=nbeats,
track_indices=[
model["state_index"]("v"),
model["state_index"]("cai"),
model["state_index"]("nai"),
],
dt=0.05,
),
2: beat.single_cell.get_steady_state(
fun=model["generalized_rush_larsen"],
init_states=model["init_state_values"](),
parameters=model["init_parameter_values"](celltype=1),
outdir=results_folder / "epi",
BCL=1000,
nbeats=nbeats,
track_indices=[model["state_index"]("v"), model["state_index"]("cai")],
dt=0.05,
),
}
# endo = 0, epi = 1, M = 2
parameters = {
0: model["init_parameter_values"](i_Stim_Amplitude=0.0, celltype=2),
1: model["init_parameter_values"](i_Stim_Amplitude=0.0, celltype=0),
2: model["init_parameter_values"](i_Stim_Amplitude=0.0, celltype=1),
}
fun = {
0: model["generalized_rush_larsen"],
1: model["generalized_rush_larsen"],
2: model["generalized_rush_larsen"],
}
v_index = {
0: model["state_index"]("v"),
1: model["state_index"]("v"),
2: model["state_index"]("v"),
}
Get steady states
time = dolfinx.fem.Constant(mesh, dolfinx.default_scalar_type(0.0))
I_s = beat.stimulation.define_stimulus(
mesh=data.mesh,
chi=chi,
time=time,
subdomain_data=data.ffun,
marker=markers["ENDO"][0],
amplitude=stim_amp,
mesh_unit=mesh_unit,
)
M = beat.conductivities.define_conductivity_tensor(
chi,
f0=data.f0,
g_il=g_il,
g_it=g_it,
g_el=g_el,
g_et=g_et,
)
params = {"preconditioner": "sor", "use_custom_preconditioner": False}
pde = beat.MonodomainModel(
time=time,
mesh=data.mesh,
M=M,
I_s=I_s,
params=params,
C_m=C_m.to(f"uF/{mesh_unit}**2").magnitude,
)
V_ode = dolfinx.fem.functionspace(data.mesh, ("P", 1))
ode = beat.odesolver.DolfinMultiODESolver(
v_ode=dolfinx.fem.Function(V_ode),
v_pde=pde.state,
markers=cfun_func,
num_states={i: len(s) for i, s in init_states.items()},
fun=fun,
init_states=init_states,
parameters=parameters,
v_index=v_index,
)
t = 0.0
solver = beat.MonodomainSplittingSolver(pde=pde, ode=ode)
vtxfname = results_folder / "slab.bp"
checkpointfname = results_folder / "slab_checkpoint.bp"
shutil.rmtree(vtxfname, ignore_errors=True)
shutil.rmtree(checkpointfname, ignore_errors=True)
vtx = dolfinx.io.VTXWriter(
comm,
vtxfname,
[solver.pde.state],
engine="BP4",
)
adios4dolfinx.write_mesh(checkpointfname, mesh)
def save(t):
v = solver.pde.state.x.array
print(f"Solve for {t=:.2f}, {v.max() =}, {v.min() =}")
vtx.write(t)
adios4dolfinx.write_function(checkpointfname, solver.pde.state, time=t, name="v")
i = 0
while t < end_time + 1e-12:
# Make sure to save at the same time steps that is used by Ambit
if i % save_freq == 0:
save(t)
solver.step((t, t + dt))
i += 1
t += dt
Solve for t=0.00, v.max() =np.float64(-89.49461598667106), v.min() =np.float64(-89.84978422825839)
Solve for t=1.00, v.max() =np.float64(96.18090213247223), v.min() =np.float64(-89.84982993061158)
Solve for t=2.00, v.max() =np.float64(156.7317581652501), v.min() =np.float64(-89.8498672815758)
Solve for t=3.00, v.max() =np.float64(60.93322179874107), v.min() =np.float64(-89.84990772868004)
Solve for t=4.00, v.max() =np.float64(40.046999771428986), v.min() =np.float64(-89.8499384314671)
Solve for t=5.00, v.max() =np.float64(30.118531218608), v.min() =np.float64(-89.8499353942576)
Solve for t=6.00, v.max() =np.float64(25.25918861495623), v.min() =np.float64(-89.84988046329886)
Solve for t=7.00, v.max() =np.float64(22.822875687583092), v.min() =np.float64(-89.8497745953058)
Solve for t=8.00, v.max() =np.float64(22.08346742786114), v.min() =np.float64(-89.84963217201073)
Solve for t=9.00, v.max() =np.float64(22.86740070084043), v.min() =np.float64(-89.84947134388037)
Solve for t=10.00, v.max() =np.float64(23.239979893675116), v.min() =np.float64(-89.84930799847754)
Solve for t=11.00, v.max() =np.float64(23.3322047847248), v.min() =np.float64(-89.84915356790438)
Solve for t=12.00, v.max() =np.float64(23.35188241485754), v.min() =np.float64(-89.84901503532018)
Solve for t=13.00, v.max() =np.float64(23.356830164032143), v.min() =np.float64(-89.84889571425754)
Solve for t=14.00, v.max() =np.float64(23.35401951196897), v.min() =np.float64(-89.84879224671721)
Solve for t=15.00, v.max() =np.float64(22.60186854921293), v.min() =np.float64(-89.84854189377153)
Solve for t=16.00, v.max() =np.float64(21.209130923616026), v.min() =np.float64(-89.84179231272114)
Solve for t=17.00, v.max() =np.float64(21.205289594434372), v.min() =np.float64(-89.56313334895438)
Solve for t=18.00, v.max() =np.float64(21.93513100869462), v.min() =np.float64(-76.95721666302443)
Solve for t=19.00, v.max() =np.float64(33.41454207029696), v.min() =np.float64(14.971675112922254)
Solve for t=20.00, v.max() =np.float64(23.465127541523454), v.min() =np.float64(14.17342600820865)
Compute conduction velocity and ECG#
threshold = 0.0
x0 = L * 0.25
x1 = L * 0.75
if mesh.geometry.dim == 2:
p_ecg = (L * 2.0, dx * 0.5)
p1 = (x0, dx * 0.5)
p2 = (x1, dx * 0.5)
else:
p_ecg = (L * 2.0, dx * 0.5, dx * 0.5) # type: ignore
p1 = (x0, dx * 0.5, dx * 0.5) # type: ignore
p2 = (x1, dx * 0.5, dx * 0.5) # type: ignore
Need to either save the functions on the input mesh using adios4dolfinx.write_function_on_input_mesh or read the mesh again see https://jsdokken.com/adios4dolfinx/docs/original_checkpoint.html
mesh = adios4dolfinx.read_mesh(comm=comm, filename=checkpointfname)
V = dolfinx.fem.functionspace(mesh, ("P", 1))
v = dolfinx.fem.Function(V)
plotter_voltage = pyvista.Plotter()
grid = pyvista.UnstructuredGrid(*dolfinx.plot.vtk_mesh(V))
grid.point_data["V"] = v.x.array
viridis = plt.get_cmap("viridis")
sargs = dict(
title_font_size=25,
label_font_size=20,
fmt="%.2e",
color="black",
position_x=0.1,
position_y=0.8,
width=0.8,
height=0.1,
)
plotter_voltage.add_mesh(
grid,
show_edges=True,
lighting=False,
cmap=viridis,
scalar_bar_args=sargs,
clim=[-90.0, 40.0],
)
times = beat.postprocess.read_timestamps(comm, checkpointfname, "v")
t1 = np.inf
t2 = np.inf
phie = []
ecg = beat.ecg.ECGRecovery(
v=v, sigma_b=1.0, C_m=C_m.to(f"uF/{mesh_unit}**2").magnitude, M=M,
)
p_ecg_form = ecg.eval(p_ecg)
gif_file = Path("voltage_slab_time.gif")
gif_file.unlink(missing_ok=True)
plotter_voltage.open_gif(gif_file.as_posix())
for t in times:
adios4dolfinx.read_function(checkpointfname, v, time=t, name="v")
ecg.solve()
phie.append(
mesh.comm.allreduce(dolfinx.fem.assemble_scalar(p_ecg_form), op=MPI.SUM),
)
grid.point_data["V"] = v.x.array
plotter_voltage.write_frame()
v1, v2 = scifem.evaluate_function(v, [p1, p2])
print(f"Read {t=:.2f}, {v1 =}, {v2 =}")
if v1 > threshold:
t1 = min(t, t1)
if v2 > threshold:
t2 = min(t, t2)
plotter_voltage.close()
Read t=0.00, v1 =array([-89.74885233]), v2 =array([-89.84978423])
Read t=1.00, v1 =array([-89.74382839]), v2 =array([-89.84285777])
Read t=2.00, v1 =array([-89.73163886]), v2 =array([-89.82798931])
Read t=3.00, v1 =array([-89.61587526]), v2 =array([-89.81826262])
Read t=4.00, v1 =array([-85.62186006]), v2 =array([-89.81229107])
Read t=5.00, v1 =array([-4.60050736]), v2 =array([-89.80853166])
Read t=6.00, v1 =array([21.08622303]), v2 =array([-89.80609628])
Read t=7.00, v1 =array([21.50192394]), v2 =array([-89.80448303])
Read t=8.00, v1 =array([21.44475013]), v2 =array([-89.80339789])
Read t=9.00, v1 =array([21.16412938]), v2 =array([-89.80266036])
Read t=10.00, v1 =array([20.81092625]), v2 =array([-89.80211725])
Read t=11.00, v1 =array([20.43555961]), v2 =array([-89.80027631])
Read t=12.00, v1 =array([20.07549438]), v2 =array([-89.74701865])
Read t=13.00, v1 =array([19.7828611]), v2 =array([-87.92275988])
Read t=14.00, v1 =array([19.60614111]), v2 =array([-32.74283281])
Read t=15.00, v1 =array([19.56103128]), v2 =array([20.28303277])
Read t=16.00, v1 =array([19.63238866]), v2 =array([19.28794502])
Read t=17.00, v1 =array([19.79060021]), v2 =array([17.80693058])
Read t=18.00, v1 =array([20.00495566]), v2 =array([16.3537289])
Read t=19.00, v1 =array([20.25124732]), v2 =array([15.11195494])
Read t=20.00, v1 =array([20.5088712]), v2 =array([14.18767015])

if not np.isclose(t1, t2):
cv = (x1 - x0) / (t2 - t1) * beat.units.ureg(f"{mesh_unit}/ms")
msg = (
f"Conduction velocity = {cv.magnitude:.3f} mm/ms or " #
f" {cv.to('m/s').magnitude:.3f} m/s or " #
f" {cv.to('cm/s').magnitude:.3f} cm/s" #
)
print(msg)
Conduction velocity = 0.056 mm/ms or 0.556 m/s or 55.556 cm/s
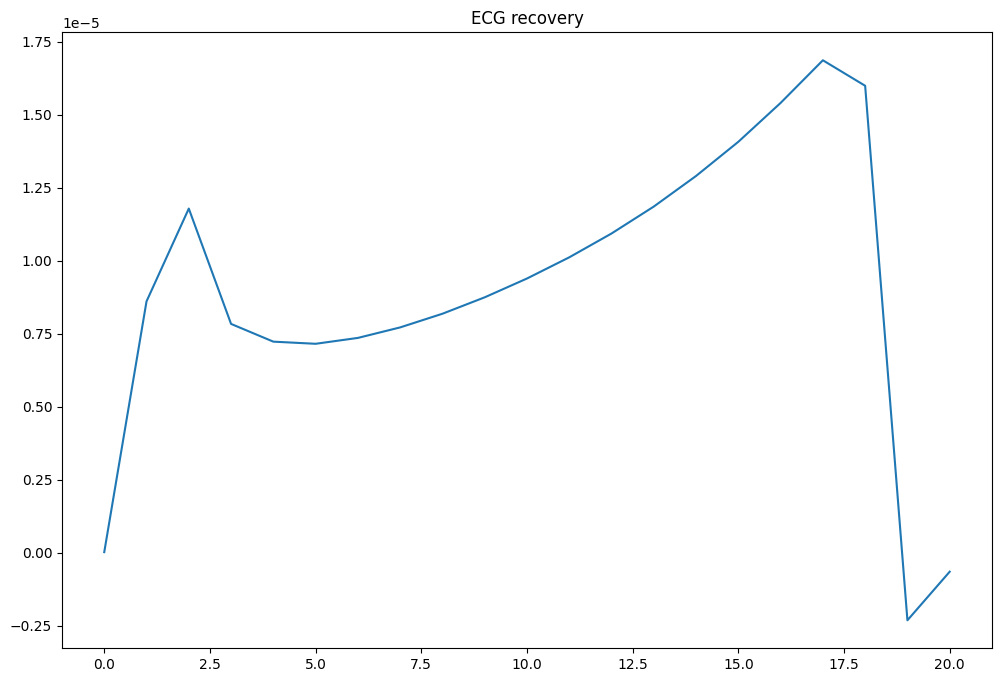
fig, ax = plt.subplots(1, 1, figsize=(12, 8))
ax.plot(times, phie)
ax.set_title("ECG recovery")
fig.savefig(results_folder / "ecg_recovery.png")